A novel approach to feed and train extra features in Seq2seq (Tensorflow & Pytorch)
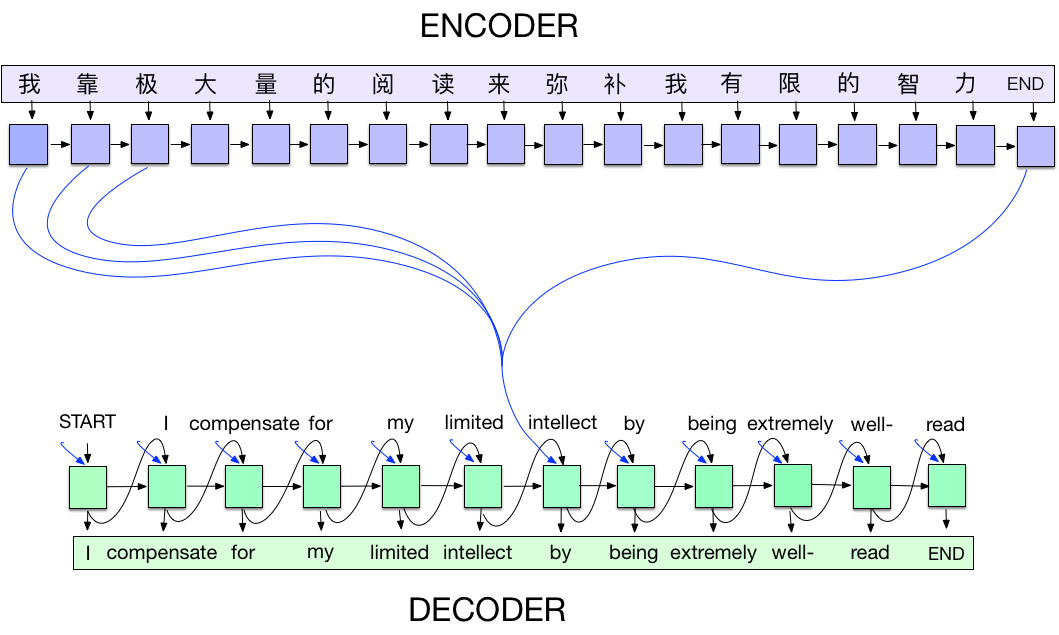
Table of Contents
- Introduction
- So what’s the problem?
- Sequence modelling before deep learning
- Data input to CRF models
- The technique
- Pseudocode
- Pytorch code
- Tf code
- Notes on the experiment
- Conclusion
Introduction
Recent developments in deep learning have given rise to a powerful alternative – discriminative models called sequence-to-sequence models, can be trained to model the conditional probability distribution of the output transcript sequence given the input sequence, directly without inverting a generative model.
Encoder-decoder neural models (Sutskever et al., 2014) are a generic deep-learning approach to sequence-to-sequence translation (Seq2Seq) tasks. Encoder Decoder network, is a model consisting of two separate RNNs called the encoder and decoder. The encoder reads an input sequence one item at a time, and outputs a vector at each step. The final output of the encoder is kept as the context vector. The decoder uses this context vector to produce a sequence of outputs one step at a time. The performance of the original sequence-to-sequence model has equal Contribution been greatly improved by the invention of soft attention, which made it possible for sequence-to sequence models to generalize better and achieve excellent results using much smaller networks on long sequences. The sequence-to-sequence model with attention had considerable empirical success on machine translation, speech recognition, image caption generation, and question answering.
If you want to quickly go through the progress of seq2seq research. Paper notes of the foundational papers in seq2seq literature are available here
To get more specific. Below are the foundational seq2seq papers listed chronologically:
Neural Machine Translation(NMT)
-
Sequence to Sequence Learning with Neural Networks[arXiv] [notes]
-
Neural Machine Translation by Jointly Learning to Align and Translate (2015)[arXiv] [notes]
-
Effective Approaches to Attention-based Neural Machine Translation [arXiv] [notes]
-
Massive Exploration of Neural Machine Translation Architectures[arXiv] [notes]
So what’s the problem ?
The training data for seq2seq involves the use of a parallel corpus (L1-L2) aligned at the level of sentences and words. Traditionally, since sequence to sequence models take only one input features at a time. Currently, there is no way we can feed more than one input feature at once to the seq2seq model.
If we consider the input data of previous statistical based approaches of sequence modelling before seq2seq. The input data on the source tokens contains extra features along with the main tokens.
For example, let’s consider the NMT problem, say I have 2 more feature columns for the corresponding source vocabulary( Feature1 here). For example, consider this below:
Feature1 Feature2 Feature3
word1 x a
word2 y b
word3 y c
.
.
So, how do we pass extra feature along with the existing word tokens as input and feed it to the encoder RNN?
Let’s consider a case where we want to train the seq2seq model for Nmt problem. Or even sequence labeling/tagging problem like a NER for that matter.
In natural language processing, it is a common task to extract words or phrases of particular types from a given sentence or paragraph. For example, when performing analysis of a corpus of news articles, we may want to know which countries are mentioned in the articles, and how many articles are related to each of these countries. This is actually a special case of sequence labelling in NLP (others include POS tagging and Chunking), in which the goal is to assign a label to each member in the sequence. In the case of identifying country names, we would like to assign a ‘country’ label to words that form part of a country name, and a ‘irrelevant’ label to all other words. For example, the following is a sentence broken down into tokens, and its desired output after the sequence labelling process:
input = ["Paris", "is", "the", "capital", "of", "France"]
output = ["I", "I", "I", "I", "I", "C"]
Say we have extra features like POS for every source token and we want a way to train those features. Instead of ignoring them and training just with the typical source-target parallel corpus training way.
To understand this a little bit more, let’s explore the statistical Sequence modelling methods before deep learning.
Sequence modelling before deep learning
i.e Statistical sequence modelling
Statistical methods have been the choice for many NLP tasks. Statistical approaches, basically utilize some machine learning methods typically supervised or semi-supervised algorithms to identify entities.
Sequence-based methods, use complete sequence of words instead of only single words or phrases. They try to predict the most likely tag for a sequence of words after being trained on a training set. Hidden Markov Model(HMM). Maximum Entropy Markov Model and Conditional Random Fields are the most common sequence-based approaches and CRFs have frequently demonstrated to be better statistical biomedical NER systems. The primary advantage of CRFs over hidden Markov models is their conditional nature, resulting in the relaxation of the independence assumptions required by HMMs in order to ensure tractable inference. CRFs outperform both MEMMs and HMMs on a number of real-world tasks in many fields, including bioinformatics, computational linguistics and speech recognition.
That said, now let us consider a specific case of how data is fed to CRF models.
Data input to CRF models
Typically in CRF models, our input file should be in the following format:
Bill CAPITALIZED noun
slept non-noun
here LOWERCASE STOPWORD non-noun
That is, each line represents one token, and has the format: feature1 feature2 … featureN label
The first column is assumed to be the token and the last column is the label. There can be other columns in the middle, which are currently not used.
The result will be a list of documents, each of which contains a list of (word, label) tuples. For example:
>>> doc[0][:10]
[('Paxar', 'N'),
('Corp', 'N'),
('said', 'I'),
('it', 'I'),
('has', 'I'),
('acquired', 'I'),
('Thermo-Print', 'N'),
Furthermore, features can be generated at run-time. Given the POS tags, we can now continue to generate more features for each of the tokens in the dataset. The features that will be useful in the training process depends on the task at hand. Below are some of the commonly used features for a word w in named entity recognition. To point a few; the words surrounding w, such as the previous and the next word or whether w is in uppercase or lowercase or number, or contains digits or contains a special character. For further notes on this, please refer this wonderful blog post.
The technique
There are few possible options to pass extra tokens to the seq2seq encoder.
- The simplest way is to combine features into a single input vector. However this only works if our RNN takes vector input, not discrete inputs (LongTensor) through an embedding layer. In that case we would want to combine the extra features after the input is embedded. Considering the features are also discrete, we would want multiple embedding layers, one for each, and combine all the results (eg. Could be an embedding of POS, or simply one-hot.). Precisely, we forward the features through the relevant embedding layers, and combine them all into one vector for the RNN.
I have sketched below illustration, to provide more clarity.
The figure below illustrates the method of using 2 different vocabulares in a single embedding matrix. Let’s say 1600 is set in W, 23 is set in F1. We can pull the vectors belonging to 2 vocabularies in single embedding matrix as shown below. (For illustration purpose; 1600, 6023 are the active tokens tokens)
Further, there is another option where in we train the network with more parameters to predict both POS and NER (only change final layers). Thus, the network would “internally” leverage the information(I haven’t tried this yet, but worth experimenting). As pointed out by the author of this blog post here.
Tensorflow seq2seq
Do the necessary changes in the file data/input_pipeline.py, data/parallel_data_provider.py, training/utils.py, models/seq2seq_model.py(update the new feature vocab here) for the the extra feature data processing.
In the file models/basic_seq2seq.py
# Get the Embedding Matrix
common_embedding = self.source_embedding
# Get the Embedding for the Main Features
source_embedded = tf.nn.embedding_lookup(common_embedding,features["source_ids"])
# Get the Embedding for Extra Feature
s1 = tf.fill(tf.shape(features["source_f1_ids"]),self.source_vocab_info.total_size)
s2 = tf.add(tf.cast(s1,tf.int64),features["source_f1_ids"])
source_f1_embedded = tf.nn.embedding_lookup(common_embedding,s2)
# combine the embedding extra feature along with the source token using vector addition
# new extended source embedding
extended_embedding = tf.add(source_embedded,source_f1_embedded)
"""
Note:
tf.fill - Creates a tensor filled with a scalar value. This operation creates a tensor of shape dims and fills it with value. (Equivalent to np.full)
tf.nn.embedding_lookup - embedding_lookup function retrieves rows(ie. the embedding vector) of the params((ie. the word ids) tensor.
# tf.add returns x + y element-wise
"""
In the above , we fill the source_f1_ids the size of the source vocabulary. And add the existing feature ids of extra feature to the size of the source vocabulary. This way we make the embedding of the extra feature to belong in the same embedding matrix.
Pytorch seq2seq
Pytorch seq2seq code
Do the necessary changes in the file nmt.py(driver code) for the extra feature data processing to pass the data path, vocabulary,etc. Likewise already did for the source tokens.
In the file Model.py
# 1/ Initialise embedding for the extra feature (input_src_f1)
def forward(self, input_src, input_trg, input_src_f1, trg_mask=None, ctx_mask=None):
# Create new embedding for the already defined src_embedding definition/initialiser
f1_emb = self.src_embedding(input_src_f1)
# Combining the source features
# Create a randomly initialised tensor of size (batch_size X sequence length, embeding_dim)
extended_embedding = Variable(torch.randn(80, len(input_src[0]), 500)).cuda()
# For every batches, pull embedding vectors of the extra feature and combine them
for i, s in enumerate(src_emb):
extended_embedding[i,:,:] = (src_emb[i,:,:] + f1_emb[i,:,:])
self.h0_encoder, self.c0_encoder = self.get_state(input_src)
src_h, (src_h_t, src_c_t) = self.encoder(
extended_embedding, (self.h0_encoder, self.c0_encoder))
"""
Note: If using only CPU, can remove .cuda()
[i,:,:] - numpy style ndarrays slicing
"""
Which is same as what we did above in tensorflow.
Source Codes
The source code for reproducing the above results can be found in the following github repository: https://github.com/iamsiva11/Seq2Seq-PyTorch
Notes on the experiment
Results and Performance
The experiment was carried out on a single modern GPU (Geforce GTX 1080, 1080ti and Tesla K80). With extended features approach a significant boost in the F1-score was inferred, close to 4-5% which was impressive.
This type of model has a large number of available hyperparameters, or knobs we can tune, all of which will affect training time and final performance. The typical training time for 100K epochs range from 6-8 hours for 1 million sentences ( Yes, the training time majorly depends on the dataset size). Hence, carrying out the experiment for large no of iterations resulted in waiting for 4-5 days to evaluate the model.
Other significant hyperparameters used in the experiment used to get higher accuracy were Maximum sequence length - 50; Bidirectional RNN as encoder ; Bahdanau Attention based Decoder; ADAM as optimiser; learning rate as 0.0001; 1 layer of encoder and 2 layers of decoder ; GRU cells were used as RNN units for both encoder and decoder; beam search methodology for decoding.
Additional notes
Also I experimented with Concatenation instead of addition by extending the embedding dimension. While getting inspired from these papers, papers and Bidirectional LSTM layers where we can concatenate (default), multiplication, average, and sum the outputs. This approach didn’t give any better results though.
Further, the vector addition operation of feature embeddings we performed earlier is a hacky way of avoiding the matrix multiplication of the embedding matrix and the one-hot vector. This way we save time and the computation cost.
Initially, I worked on pytorch for this project, but got to know their CPU backend is relatively slow resulting in slow predictions. And I switched to tensorflow after that. Even though I had worked in tf previously in its early days on lot of computer vision problems. I was impressed with the rapid improvement in the APIs. Providing better control while giving an option to switch between low level and high level apis depending on our needs; as this was not the case before v1.0. Much impressed with tensorflow.
Moreover, I like pytorch for quick experimentation and research purposes though. As getting production ready has not been their focus from first. That said they are good at what they do. While tensorflow’s power comes with a bit of verbosity. Which is nothing to complain about, if we look at the scale and big picture of things it can help us accomplish in the long term.
Conclusion
I would like to thank the google brain team for open sourcing the seq2seq tensorflow code, @spro on his valuable inputs for handling this problem, @MaximumEntropy for the pytorch seq2seq code.
References
Xuezhe Ma and Eduard Hovy, “End-to-end Sequence Labeling via Bi-directional LSTM-CNNs-CRF”
Lafferty, J., McCallum, A., Pereira, F. (2001). “Conditional random fields: Probabilistic models for segmenting and labeling sequence data”. Proc. 18th International Conf. on Machine Learning. Morgan Kaufmann. pp. 282–289.
Erdogan, H. (2010). Sequence Labeling: Generative and Discriminative Approaches - Hidden Markov Models, Conditional Random Fields and Structured SVMs. ICMLA 2010 Tutorial.


